Are Prokaryotic Cells In Humans

Diagram of a typical prokaryotic cell
A prokaryote () is a single-celled organism that lacks a nucleus and other membrane-bound organelles.[1] The word prokaryote comes from the Greek πρό ( pro , 'earlier') and κάρυον ( karyon , 'nut' or 'kernel').[2] [3] In the two-empire organization arising from the piece of work of Édouard Chatton, prokaryotes were classified inside the empire Prokaryota.[4] But in the 3-domain system, based upon molecular analysis, prokaryotes are divided into ii domains: Bacteria (formerly Eubacteria) and Archaea (formerly Archaebacteria). Organisms with nuclei are placed in a third domain, Eukaryota.[v] In the report of the origins of life, prokaryotes are thought to have arisen before eukaryotes.
Besides the absence of a nucleus, prokaryotes also lack mitochondria, or near of the other membrane-bound organelles that characterize the eukaryotic cell. It was once thought that prokaryotic cellular components within the cytoplasm were unenclosed, except for an outer cell membrane, but bacterial microcompartments, which are thought to be simple organelles enclosed in protein shells, have been discovered,[half dozen] [7] forth with other prokaryotic organelles.[viii] While being unicellular, some prokaryotes, such every bit cyanobacteria, may form large colonies. Others, such as myxobacteria, have multicellular stages in their life cycles.[ix] Prokaryotes are asexual, reproducing without fusion of gametes, although horizontal gene transfer likewise takes identify.
Molecular studies accept provided insight into the evolution and interrelationships of the iii domains of life.[10] The division between prokaryotes and eukaryotes reflects the existence of two very different levels of cellular organisation; only eukaryotic cells have an enveloped nucleus that contains its chromosomal DNA, and other feature membrane-leap organelles including mitochondria. Distinctive types of prokaryotes include extremophiles and methanogens; these are common in some extreme environments.[1]
History [edit]
The stardom betwixt prokaryotes and eukaryotes was firmly established by the microbiologists Roger Stanier and C. B. van Niel in their 1962 paper The concept of a bacterium [xi] (though spelled procaryote and eucaryote in that location). That paper cites Édouard Chatton's 1937 book Titres et Travaux Scientifiques [12] for using those terms and recognizing the distinction. Ane reason for this nomenclature was so that what was then often called blue-green algae (now called cyanobacteria) would not be classified equally plants but grouped with bacteria.
Structure [edit]
Prokaryotes take a prokaryotic cytoskeleton that is more primitive than that of the eukaryotes. Also homologues of actin and tubulin (MreB and FtsZ), the helically bundled building-block of the flagellum, flagellin, is one of the near significant cytoskeletal proteins of bacteria, as it provides structural backgrounds of chemotaxis, the basic cell physiological response of bacteria. At least some prokaryotes too contain intracellular structures that can be seen equally primitive organelles. Membranous organelles (or intracellular membranes) are known in some groups of prokaryotes, such every bit vacuoles or membrane systems devoted to special metabolic properties, such as photosynthesis or chemolithotrophy. In improver, some species besides comprise saccharide-enclosed microcompartments, which have distinct physiological roles (e.one thousand. carboxysomes or gas vacuoles).
About prokaryotes are between 1 µm and ten µm, merely they tin can vary in size from 0.2 µm (Mycoplasma genitalium) to 750 µm (Thiomargarita namibiensis).
| Prokaryotic prison cell structure | Clarification |
|---|---|
| Flagellum (not always present) | Long, whip-like protrusion that aids cellular locomotion used past both gram positive and gram negative organisms. |
| Jail cell membrane | Surrounds the cell's cytoplasm and regulates the flow of substances in and out of the prison cell. |
| Cell wall (except genera Mycoplasma and Thermoplasma) | Outer covering of most cells that protects the bacterial cell and gives it shape. |
| Cytoplasm | A gel-like substance composed mainly of h2o that besides contains enzymes, salts, cell components, and various organic molecules. |
| Ribosome | Cell structures responsible for poly peptide product. |
| Nucleoid | Area of the cytoplasm that contains the prokaryote's single DNA molecule. |
| Glycocalyx (merely in some types of prokaryotes) | A glycoprotein-polysaccharide roofing that surrounds the cell membranes. |
| Cytoplasmic inclusions | They contain the inclusion bodies like ribosomes and larger masses scattered in the cytoplasmic matrix. |
Morphology [edit]
Prokaryotic cells have various shapes; the four basic shapes of bacteria are:[thirteen]
- Cocci – A bacterium that is spherical or ovoid is chosen a coccus (Plural, cocci). due east.g. Streptococcus, Staphylococcus.
- Bacilli – A bacterium with cylindrical shape called rod or a bacillus (Plural, bacilli).
- Spiral leaner – Some rods twist into spiral shapes and are chosen spirilla (singular, spirillum).
- Vibrio – comma-shaped
The archaeon Haloquadratum has flat foursquare-shaped cells.[fourteen]
Reproduction [edit]
Bacteria and archaea reproduce through asexual reproduction, normally by binary fission. Genetic exchange and recombination all the same occur, but this is a form of horizontal gene transfer and is not a replicative process, only involving the transference of Deoxyribonucleic acid between two cells, as in bacterial conjugation.
DNA transfer [edit]
DNA transfer between prokaryotic cells occurs in bacteria and archaea, although it has been mainly studied in bacteria. In bacteria, cistron transfer occurs past three processes. These are (1) bacterial virus (bacteriophage)-mediated transduction, (ii) plasmid-mediated conjugation, and (three) natural transformation. Transduction of bacterial genes past bacteriophage appears to reflect an occasional error during intracellular assembly of virus particles, rather than an adaptation of the host bacteria. The transfer of bacterial Deoxyribonucleic acid is under the control of the bacteriophage'southward genes rather than bacterial genes. Conjugation in the well-studied E. coli system is controlled by plasmid genes, and is an accommodation for distributing copies of a plasmid from one bacterial host to another. Infrequently during this process, a plasmid may integrate into the host bacterial chromosome, and subsequently transfer role of the host bacterial DNA to another bacterium. Plasmid mediated transfer of host bacterial Deoxyribonucleic acid (conjugation) also appears to be an accidental procedure rather than a bacterial adaptation.
3D animation of a prokaryotic cell that shows all the elements that information technology is composed of
Natural bacterial transformation involves the transfer of Deoxyribonucleic acid from one bacterium to another through the intervening medium. Unlike transduction and conjugation, transformation is clearly a bacterial accommodation for DNA transfer, because it depends on numerous bacterial cistron products that specifically collaborate to perform this complex process.[15] For a bacterium to bind, take up and recombine donor Dna into its own chromosome, information technology must first enter a special physiological state chosen competence. About 40 genes are required in Bacillus subtilis for the development of competence.[sixteen] The length of Dna transferred during B. subtilis transformation can exist as much as a tertiary to the whole chromosome.[17] [18] Transformation is a common mode of DNA transfer, and 67 prokaryotic species are thus far known to be naturally competent for transformation.[19]
Among archaea, Halobacterium volcanii forms cytoplasmic bridges between cells that appear to be used for transfer of Deoxyribonucleic acid from one prison cell to another.[20] Another archaeon, Sulfolobus solfataricus, transfers DNA betwixt cells by direct contact. Frols et al.[21] found that exposure of South. solfataricus to Dna damaging agents induces cellular aggregation, and suggested that cellular aggregation may enhance Dna transfer among cells to provide increased repair of damaged DNA via homologous recombination.
[edit]
While prokaryotes are considered strictly unicellular, most can course stable aggregate communities.[22] When such communities are encased in a stabilizing polymer matrix ("slime"), they may exist chosen "biofilms".[23] Cells in biofilms often testify distinct patterns of gene expression (phenotypic differentiation) in time and space. Also, as with multicellular eukaryotes, these changes in expression often announced to result from cell-to-cell signaling, a miracle known as quorum sensing.
Biofilms may be highly heterogeneous and structurally complex and may adhere to solid surfaces, or be at liquid-air interfaces, or potentially even liquid-liquid interfaces. Bacterial biofilms are often made up of microcolonies (approximately dome-shaped masses of leaner and matrix) separated by "voids" through which the medium (due east.chiliad., water) may catamenia easily. The microcolonies may bring together together above the substratum to form a continuous layer, closing the network of channels separating microcolonies. This structural complexity—combined with observations that oxygen limitation (a ubiquitous challenge for anything growing in size beyond the scale of diffusion) is at least partially eased by movement of medium throughout the biofilm—has led some to speculate that this may constitute a circulatory arrangement[24] and many researchers take started calling prokaryotic communities multicellular (for example [25]). Differential jail cell expression, collective behavior, signaling, programmed cell death, and (in some cases) discrete biological dispersal[26] events all seem to point in this direction. However, these colonies are seldom if ever founded by a single founder (in the mode that animals and plants are founded past single cells), which presents a number of theoretical issues. Most explanations of co-operation and the evolution of multicellularity have focused on high relatedness between members of a group (or colony, or whole organism). If a copy of a gene is present in all members of a group, behaviors that promote cooperation between members may allow those members to have (on average) greater fitness than a similar grouping of selfish individuals[27] (see inclusive fettle and Hamilton'due south rule).
Should these instances of prokaryotic sociality testify to exist the rule rather than the exception, information technology would have serious implications for the fashion we view prokaryotes in general, and the way we deal with them in medicine.[28] Bacterial biofilms may exist 100 times more resistant to antibiotics than free-living unicells and may be nearly impossible to remove from surfaces once they have colonized them.[29] Other aspects of bacterial cooperation—such as bacterial conjugation and quorum-sensing-mediated pathogenicity, present boosted challenges to researchers and medical professionals seeking to treat the associated diseases.
Environment [edit]

Phylogenetic ring showing the diversity of prokaryotes, and symbiogenetic origins of eukaryotes
Prokaryotes have diversified profoundly throughout their long being. The metabolism of prokaryotes is far more than varied than that of eukaryotes, leading to many highly distinct prokaryotic types. For example, in addition to using photosynthesis or organic compounds for energy, as eukaryotes do, prokaryotes may obtain energy from inorganic compounds such as hydrogen sulfide. This enables prokaryotes to thrive in harsh environments as cold equally the snowfall surface of Antarctica, studied in cryobiology, or every bit hot as undersea hydrothermal vents and land-based hot springs.
Prokaryotes live in nearly all environments on Earth. Some archaea and bacteria are extremophiles, thriving in harsh atmospheric condition, such as high temperatures (thermophiles) or loftier salinity (halophiles).[30] Many archaea grow as plankton in the oceans. Symbiotic prokaryotes live in or on the bodies of other organisms, including humans. Prokaryote take high populations in the soil - including the rhizosphere and rhizosheath. Soil prokaryotes are notwithstanding heavily undercharacterized despite their easy proximity to humans and their tremendous economic importance to agriculture.[31]

Classification [edit]
In 1977, Carl Woese proposed dividing prokaryotes into the Bacteria and Archaea (originally Eubacteria and Archaebacteria) considering of the major differences in the structure and genetics between the two groups of organisms. Archaea were originally idea to be extremophiles, living but in inhospitable conditions such equally extremes of temperature, pH, and radiation but take since been plant in all types of habitats. The resulting system of Eukaryota (also chosen "Eucarya"), Bacteria, and Archaea is called the iii-domain system, replacing the traditional two-empire arrangement.[32] [33]
Phylogenetic tree [edit]
According to the phylogenetic analysis of Zhu (2019), the relationships could be the following:[34]
Evolution [edit]

Diagram of the origin of life with the Eukaryotes appearing early, not derived from Prokaryotes, as proposed by Richard Egel in 2012. This view, 1 of many on the relative positions of Prokaryotes and Eukaryotes, implies that the universal common ancestor was relatively big and complex.[35]
A widespread current model of the evolution of the first living organisms is that these were some class of prokaryotes, which may have evolved out of protocells, while the eukaryotes evolved after in the history of life.[36] Some authors have questioned this conclusion, arguing that the current set of prokaryotic species may have evolved from more complex eukaryotic ancestors through a process of simplification.[37] [38] [39] Others have argued that the three domains of life arose simultaneously, from a set up of varied cells that formed a single gene pool.[twoscore] This controversy was summarized in 2005:[41]
There is no consensus among biologists concerning the position of the eukaryotes in the overall scheme of cell evolution. Electric current opinions on the origin and position of eukaryotes bridge a wide spectrum including the views that eukaryotes arose first in evolution and that prokaryotes descend from them, that eukaryotes arose contemporaneously with eubacteria and archaebacteria and hence represent a chief line of descent of equal age and rank every bit the prokaryotes, that eukaryotes arose through a symbiotic issue entailing an endosymbiotic origin of the nucleus, that eukaryotes arose without endosymbiosis, and that eukaryotes arose through a symbiotic event entailing a simultaneous endosymbiotic origin of the flagellum and the nucleus, in addition to many other models, which have been reviewed and summarized elsewhere.
The oldest known fossilized prokaryotes were laid downwardly approximately 3.5 billion years ago, only almost one billion years after the formation of the Earth's crust. Eukaryotes but appear in the fossil record after, and may accept formed from endosymbiosis of multiple prokaryote ancestors. The oldest known fossil eukaryotes are about one.7 billion years sometime. However, some genetic evidence suggests eukaryotes appeared as early on as 3 billion years agone.[42]
While World is the only place in the universe where life is known to exist, some have suggested that at that place is evidence on Mars of fossil or living prokaryotes.[43] [44] Even so, this possibility remains the bailiwick of considerable debate and skepticism.[45] [46]
Relationship to eukaryotes [edit]

Comparison of eukaryotes vs. prokaryotes
The sectionalisation between prokaryotes and eukaryotes is usually considered the almost of import distinction or difference amongst organisms. The distinction is that eukaryotic cells have a "true" nucleus containing their DNA, whereas prokaryotic cells practise non have a nucleus.
Both eukaryotes and prokaryotes contain large RNA/protein structures chosen ribosomes, which produce poly peptide, but the ribosomes of prokaryotes are smaller than those of eukaryotes. Mitochondria and chloroplasts, two organelles constitute in many eukaryotic cells, comprise ribosomes similar in size and makeup to those plant in prokaryotes.[47] This is one of many pieces of bear witness that mitochondria and chloroplasts are descended from complimentary-living leaner. The endosymbiotic theory holds that early eukaryotic cells took in primitive prokaryotic cells by phagocytosis and adapted themselves to incorporate their structures, leading to the mitochondria and chloroplasts.
The genome in a prokaryote is held within a Dna/poly peptide circuitous in the cytosol chosen the nucleoid, which lacks a nuclear envelope.[48] The complex contains a single, cyclic, double-stranded molecule of stable chromosomal DNA, in contrast to the multiple linear, compact, highly organized chromosomes found in eukaryotic cells. In addition, many important genes of prokaryotes are stored in separate circular DNA structures called plasmids.[ii] Like Eukaryotes, prokaryotes may partially duplicate genetic cloth, and tin accept a haploid chromosomal composition that is partially replicated, a status known as merodiploidy.[49]
Prokaryotes lack mitochondria and chloroplasts. Instead, processes such as oxidative phosphorylation and photosynthesis accept place beyond the prokaryotic jail cell membrane.[l] Nevertheless, prokaryotes exercise possess some internal structures, such as prokaryotic cytoskeletons.[51] [52] It has been suggested that the bacterial phylum Planctomycetota has a membrane around the nucleoid and contains other membrane-jump cellular structures.[53] However, farther investigation revealed that Planctomycetota cells are not compartmentalized or nucleated and, like other bacterial membrane systems, are interconnected.[54]
Prokaryotic cells are unremarkably much smaller than eukaryotic cells.[2] Therefore, prokaryotes accept a larger surface-surface area-to-volume ratio, giving them a higher metabolic charge per unit, a higher growth rate, and as a consequence, a shorter generation time than eukaryotes.[2]
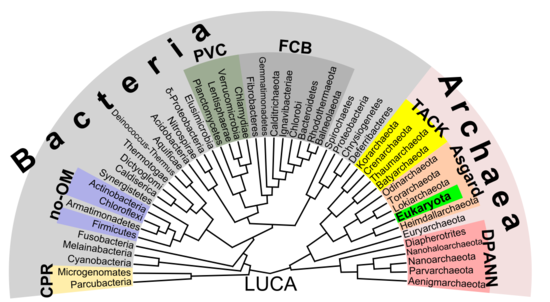
Phylogenetic tree showing the diversity of prokaryotes.[55] This 2018 proposal shows eukaryotes emerging from the archaean Asgard grouping which represents a modern version of the eocyte hypothesis. Unlike earlier assumptions, the division between bacteria and the residuum is the nigh of import divergence between organisms.
In that location is increasing evidence that the roots of the eukaryotes are to be found in (or at least next to) the archaean asgard group, perhaps Heimdallarchaeota (an idea which is a modern version of the 1984 eocyte hypothesis, eocytes existence an old synonym for Thermoproteota, a taxon to be found nearby the then-unknown Asgard group)[55] For example, histones which usually package Dna in eukaryotic nuclei, have also been institute in several archaean groups, giving evidence for homology. This thought might clarify the mysterious predecessor of eukaryotic cells (eucytes) which engulfed an alphaproteobacterium forming the beginning eucyte (LECA, last eastwardukaryotic common ancestor) according to endosymbiotic theory. In that location might have been some boosted back up by viruses, called viral eukaryogenesis. The non-bacterial group comprising archaea and eukaryota was called Neomura by Thomas Cavalier-Smith in 2002.[56] Nonetheless, in a cladistic view, eukaryota are archaea in the same sense as birds are dinosaurs because they evolved from the maniraptora dinosaur group. In dissimilarity, archaea without eukaryota appear to be a paraphyletic group, just like dinosaurs without birds.
Prokaryotes may be divide into 2 groups [edit]
Unlike the above supposition of a fundamental carve up between prokaryotes and eukaryotes, the most important difference between biota may be the sectionalization betwixt bacteria and the residuum (archaea and eukaryota).[55] For example, DNA replication differs fundamentally between bacteria and archaea (including that in eukaryotic nuclei), and it may not exist homologous between these two groups.[57] Moreover, ATP synthase, though common (homologous) in all organisms, differs greatly between leaner (including eukaryotic organelles such as mitochondria and chloroplasts) and the archaea/eukaryote nucleus grouping. The final common antecessor of all life (chosen LUCA, last universal common ancestor) should accept possessed an early version of this protein complex. Every bit ATP synthase is obligate membrane bound, this supports the supposition that LUCA was a cellular organism. The RNA world hypothesis might analyze this scenario, as LUCA might take been a ribocyte (besides called ribocell) lacking Dna, but with an RNA genome built by ribosomes as primordial self-replicating entities.[58] A Peptide-RNA earth (also called RNP world) hypothesis has been proposed based on the thought that oligopeptides may have been congenital together with primordial nucleic acids at the same fourth dimension, which too supports the concept of a ribocyte as LUCA. The feature of Deoxyribonucleic acid equally the material base of the genome might take so been adopted separately in bacteria and in archaea (and afterward eukaryote nuclei), presumably past help of some viruses (perhaps retroviruses as they could reverse transcribe RNA to Deoxyribonucleic acid).[59] As a result, prokaryota comprising bacteria and archaea may too be polyphyletic.
See likewise [edit]
- Actinonin
- Bacterial prison cell construction
- Combrex
- Evolution of cells
- Development of sexual reproduction
- Listing of sequenced archaeal genomes
- List of sequenced bacterial genomes
- Marine prokaryotes
- Monera, an obsolete kingdom including Archaea and Leaner
- Nanobacterium
- Nanobe
- Parakaryon myojinensis
- ProGlycProt
References [edit]
- ^ a b NC State University. "Prokaryotes: Single-celled Organisms".
- ^ a b c d Campbell, N. "Biology:Concepts & Connections". Pearson Education. San Francisco: 2003.
- ^ "prokaryote". Online Etymology Dictionary.
- ^ Sapp, J. (2005). "The Prokaryote-Eukaryote Dichotomy: Meanings and Mythology". Microbiology and Molecular Biology Reviews. 69 (two): 292–305. doi:10.1128/MMBR.69.2.292-305.2005. PMC1197417. PMID 15944457.
- ^ Coté K, De Tullio M (2010). "Beyond Prokaryotes and Eukaryotes: Planctomycetes and Cell Organization". Nature.
- ^ Kerfeld CA, Sawaya MR, Tanaka Due south, Nguyen CV, Phillips Chiliad, Beeby M, Yeates TO (August 2005). "Protein structures forming the beat of primitive bacterial organelles". Scientific discipline. 309 (5736): 936–eight. Bibcode:2005Sci...309..936K. CiteSeerXten.one.1.1026.896. doi:10.1126/science.1113397. PMID 16081736. S2CID 24561197.
- ^ Murat D, Byrne M, Komeili A (October 2010). "Cell biological science of prokaryotic organelles". Cold Spring Harbor Perspectives in Biology. 2 (10): a000422. doi:ten.1101/cshperspect.a000422. PMC2944366. PMID 20739411.
- ^ Murat, Dorothee; Byrne, Meghan; Komeili, Arash (2010-10-01). "Cell Biology of Prokaryotic Organelles". Cold Spring Harbor Perspectives in Biology. 2 (10): a000422. doi:10.1101/cshperspect.a000422. PMC2944366. PMID 20739411.
- ^ Kaiser D (October 2003). "Coupling cell movement to multicellular development in myxobacteria". Nature Reviews. Microbiology. i (i): 45–54. doi:ten.1038/nrmicro733. PMID 15040179. S2CID 9486133.
- ^ Sung KH, Song HK (July 22, 2014). "Insights into the molecular evolution of HslU ATPase through biochemical and mutational analyses". PLOS ONE. ix (7): e103027. Bibcode:2014PLoSO...9j3027S. doi:x.1371/periodical.pone.0103027. PMC4106860. PMID 25050622.
- ^ Stanier RY, Van Niel CB (1962). "The concept of a bacterium". Archiv für Mikrobiologie. 42 (1): 17–35. doi:10.1007/BF00425185. PMID 13916221. S2CID 29859498.
- ^ Chatton É (1937). Titres Et Travaux Scientifiques (1906-1937) De Edouard Chatton. Sète: Impr. E. Sottano.
- ^ Bauman RW, Tizard IR, Machunis-Masouka Eastward (2006). Microbiology . San Francisco: Pearson Benjamin Cummings. ISBN978-0-8053-7693-vii.
- ^ Stoeckenius West (October 1981). "Walsby's square bacterium: fine structure of an orthogonal procaryote". Journal of Bacteriology. 148 (1): 352–60. doi:x.1128/JB.148.one.352-360.1981. PMC216199. PMID 7287626.
- ^ Chen I, Dubnau D (March 2004). "Deoxyribonucleic acid uptake during bacterial transformation". Nature Reviews. Microbiology. 2 (iii): 241–9. doi:10.1038/nrmicro844. PMID 15083159. S2CID 205499369.
- ^ Solomon JM, Grossman Ad (April 1996). "Who's competent and when: regulation of natural genetic competence in bacteria". Trends in Genetics. 12 (4): 150–5. doi:10.1016/0168-9525(96)10014-7. PMID 8901420.
- ^ Akamatsu T, Taguchi H (Apr 2001). "Incorporation of the whole chromosomal DNA in protoplast lysates into competent cells of Bacillus subtilis". Bioscience, Biotechnology, and Biochemistry. 65 (4): 823–9. doi:ten.1271/bbb.65.823. PMID 11388459. S2CID 30118947.
- ^ Saito Y, Taguchi H, Akamatsu T (March 2006). "Fate of transforming bacterial genome post-obit incorporation into competent cells of Bacillus subtilis: a continuous length of incorporated Dna". Journal of Bioscience and Bioengineering. 101 (3): 257–62. doi:x.1263/jbb.101.257. PMID 16716928.
- ^ Johnsborg O, Eldholm 5, Håvarstein LS (December 2007). "Natural genetic transformation: prevalence, mechanisms and function". Enquiry in Microbiology. 158 (10): 767–78. doi:10.1016/j.resmic.2007.09.004. PMID 17997281.
- ^ Rosenshine I, Tchelet R, Mevarech K (September 1989). "The mechanism of DNA transfer in the mating system of an archaebacterium". Science. 245 (4924): 1387–nine. Bibcode:1989Sci...245.1387R. doi:10.1126/science.2818746. PMID 2818746.
- ^ Fröls Southward, Ajon M, Wagner Chiliad, Teichmann D, Zolghadr B, Folea One thousand, Boekema EJ, Driessen AJ, Schleper C, Albers SV (November 2008). "UV-inducible cellular aggregation of the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by pili formation" (PDF). Molecular Microbiology. 70 (iv): 938–52. doi:10.1111/j.1365-2958.2008.06459.10. PMID 18990182. S2CID 12797510.
- ^ Madigan T (2012). Brock biology of microorganisms (13th ed.). San Francisco: Benjamin Cummings. ISBN9780321649638.
- ^ Costerton JW (2007). "Directly Observations". The Biofilm Primer. Springer Series on Biofilms. Vol. i. Berlin, Heidelberg: Springer. pp. three–iv. doi:10.1007/978-iii-540-68022-2_2. ISBN978-3-540-68021-5.
- ^ Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM (October 1995). "Microbial biofilms". Annual Review of Microbiology. 49 (ane): 711–45. doi:10.1146/annurev.mi.49.100195.003431. PMID 8561477.
- ^ Shapiro JA (1998). "Thinking about bacterial populations as multicellular organisms" (PDF). Almanac Review of Microbiology. 52 (1): 81–104. doi:10.1146/annurev.micro.52.one.81. PMID 9891794. Archived from the original (PDF) on 2011-07-17.
- ^ Chua SL, Liu Y, Yam JK, Chen Y, Vejborg RM, Tan BG, Kjelleberg S, Tolker-Nielsen T, Givskov Thousand, Yang L (July 2014). "Dispersed cells represent a distinct phase in the transition from bacterial biofilm to planktonic lifestyles". Nature Communications. 5 (1): 4462. Bibcode:2014NatCo...5.4462C. doi:10.1038/ncomms5462. PMID 25042103.
- ^ Hamilton WD (July 1964). "The genetical evolution of social behaviour. 2". Journal of Theoretical Biology. seven (1): 17–52. Bibcode:1964JThBi...7...17H. doi:ten.1016/0022-5193(64)90039-6. PMID 5875340.
- ^ Balaban N, Ren D, Givskov M, Rasmussen TB (2008). "Introduction". Control of Biofilm Infections by Bespeak Manipulation. Springer Series on Biofilms. Vol. 2. Berlin, Heidelberg: Springer. pp. 1–eleven. doi:ten.1007/7142_2007_006. ISBN978-three-540-73852-7.
- ^ Costerton JW, Stewart PS, Greenberg EP (May 1999). "Bacterial biofilms: a common cause of persistent infections". Science. 284 (5418): 1318–22. Bibcode:1999Sci...284.1318C. doi:x.1126/scientific discipline.284.5418.1318. PMID 10334980. S2CID 27364291.
- ^ Hogan CM (2010). "Extremophile". In Monosson E, Cleveland C (eds.). Encyclopedia of Earth. National Quango of Science & the Surroundings.
- ^ Cobián Güemes, Ana Georgina; Youle, Merry; Cantú, Vito Adrian; Felts, Ben; Nulton, James; Rohwer, Forest (2016-09-29). "Viruses every bit Winners in the Game of Life". Almanac Review of Virology. Annual Reviews. 3 (one): 197–214. doi:10.1146/annurev-virology-100114-054952. ISSN 2327-056X. PMID 27741409. S2CID 36517589.
- ^ Woese CR (March 1994). "At that place must be a prokaryote somewhere: microbiology's search for itself". Microbiological Reviews. 58 (ane): 1–9. doi:ten.1128/MMBR.58.1.ane-ix.1994. PMC372949. PMID 8177167.
- ^ Sapp J (June 2005). "The prokaryote-eukaryote dichotomy: meanings and mythology". Microbiology and Molecular Biology Reviews. 69 (two): 292–305. doi:ten.1128/MMBR.69.two.292-305.2005. PMC1197417. PMID 15944457.
- ^ Zhu, Qiyun; Mai, Uyen; Pfeiffer, Wayne; Janssen, Stefan; Asnicar, Francesco; Sanders, Jon G.; Belda-Ferre, Pedro; Al-Ghalith, Gabriel A.; Kopylova, Evguenia; McDonald, Daniel; Kosciolek, Tomasz; Yin, John B.; Huang, Shi; Salam, Nimaichand; Jiao, Jian-Yu; Wu, Zijun; Xu, Zhenjiang Z.; Cantrell, Kalen; Yang, Yimeng; Sayyari, Erfan; Rabiee, Maryam; Morton, James T.; Podell, Sheila; Knights, Dan; Li, Wen-Jun; Huttenhower, Curtis; Segata, Nicola; Smarr, Larry; Mirarab, Siavash; Knight, Rob (2019). "Phylogenomics of 10,575 genomes reveals evolutionary proximity between domains Bacteria and Archaea". Nature Communications. 10 (i): 5477. Bibcode:2019NatCo..ten.5477Z. doi:10.1038/s41467-019-13443-iv. PMC6889312. PMID 31792218.
- ^ Egel R (January 2012). "Primal eukaryogenesis: on the communal nature of precellular States, ancestral to modern life". Life. 2 (1): 170–212. doi:ten.3390/life2010170. PMC4187143. PMID 25382122.
- ^ Zimmer C (August 2009). "Origins. On the origin of eukaryotes". Science. 325 (5941): 666–8. doi:10.1126/science.325_666. PMID 19661396.
- ^ Brown JR (February 2003). "Ancient horizontal gene transfer". Nature Reviews. Genetics. iv (two): 121–32. doi:10.1038/nrg1000. PMID 12560809. S2CID 22294114.
- ^ Forterre P, Philippe H (October 1999). "Where is the root of the universal tree of life?". BioEssays. 21 (10): 871–9. doi:10.1002/(SICI)1521-1878(199910)21:10<871::AID-BIES10>three.0.CO;2-Q. PMID 10497338.
- ^ Poole A, Jeffares D, Penny D (October 1999). "Early evolution: prokaryotes, the new kids on the block". BioEssays. 21 (10): 880–9. doi:x.1002/(SICI)1521-1878(199910)21:10<880::AID-BIES11>iii.0.CO;2-P. PMID 10497339. S2CID 45607498.
- ^ Woese C (June 1998). "The universal ancestor". Proceedings of the National Academy of Sciences of the United states of america of America. 95 (12): 6854–9. Bibcode:1998PNAS...95.6854W. doi:x.1073/pnas.95.12.6854. PMC22660. PMID 9618502.
- ^ Martin, William. Woe is the Tree of Life. In Microbial Phylogeny and Development: Concepts and Controversies (ed. January Sapp). Oxford: Oxford University Press; 2005: 139.
- ^ Carl Woese, J Peter Gogarten, "When did eukaryotic cells (cells with nuclei and other internal organelles) first evolve? What do nosotros know about how they evolved from before life-forms?" Scientific American, October 21, 1999.
- ^ McSween HY (July 1997). "Evidence for life in a martian meteorite?". GSA Today. 7 (7): 1–7. PMID 11541665.
- ^ McKay DS, Gibson EK, Thomas-Keprta KL, Vali H, Romanek CS, Clemett SJ, Chillier XD, Maechling CR, Zare RN (August 1996). "Search for by life on Mars: possible relic biogenic activity in martian meteorite ALH84001". Science. 273 (5277): 924–30. Bibcode:1996Sci...273..924M. doi:10.1126/science.273.5277.924. PMID 8688069. S2CID 40690489.
- ^ Crenson M (2006-08-06). "After 10 years, few believe life on Mars". Associated Press (on space.com]). Archived from the original on 2006-08-09. Retrieved 2006-08-06 .
- ^ Scott ER (Feb 1999). "Origin of carbonate-magnetite-sulfide assemblages in Martian meteorite ALH84001". Journal of Geophysical Research. 104 (E2): 3803–thirteen. Bibcode:1999JGR...104.3803S. doi:ten.1029/1998JE900034. PMID 11542931.
- ^ Bruce Alberts; et al. (2002). The Molecular Biological science of the Cell (4th ed.). Garland Science. p. 808. ISBN0-8153-3218-ane.
- ^ Thanbichler Thou, Wang SC, Shapiro L (October 2005). "The bacterial nucleoid: a highly organized and dynamic structure". Journal of Cellular Biochemistry. 96 (3): 506–21. doi:10.1002/jcb.20519. PMID 15988757. S2CID 25355087.
- ^ Johnston C, Caymaris South, Zomer A, Bootsma HJ, Prudhomme G, Granadel C, Hermans PW, Polard P, Martin B, Claverys JP (2013). "Natural genetic transformation generates a population of merodiploids in Streptococcus pneumoniae". PLOS Genetics. 9 (9): e1003819. doi:ten.1371/journal.pgen.1003819. PMC3784515. PMID 24086154.
- ^ Harold FM (June 1972). "Conservation and transformation of free energy by bacterial membranes". Bacteriological Reviews. 36 (ii): 172–230. doi:10.1128/MMBR.36.2.172-230.1972. PMC408323. PMID 4261111.
- ^ Shih YL, Rothfield L (September 2006). "The bacterial cytoskeleton". Microbiology and Molecular Biological science Reviews. 70 (3): 729–54. doi:x.1128/MMBR.00017-06. PMC1594594. PMID 16959967.
- ^ Michie KA, Löwe J (2006). "Dynamic filaments of the bacterial cytoskeleton" (PDF). Almanac Review of Biochemistry. 75 (1): 467–92. doi:ten.1146/annurev.biochem.75.103004.142452. PMID 16756499. Archived from the original (PDF) on November 17, 2006.
- ^ Fuerst JA (2005). "Intracellular compartmentation in planctomycetes". Annual Review of Microbiology. 59 (1): 299–328. doi:10.1146/annurev.micro.59.030804.121258. PMID 15910279.
- ^ Santarella-Mellwig R, Pruggnaller S, Roos N, Mattaj IW, Devos DP (2013). "Three-dimensional reconstruction of leaner with a complex endomembrane organization". PLOS Biology. 11 (5): e1001565. doi:10.1371/journal.pbio.1001565. PMC3660258. PMID 23700385.
- ^ a b c Castelle CJ, Banfield JF (March 2018). "Major New Microbial Groups Expand Variety and Alter our Agreement of the Tree of Life". Jail cell. 172 (6): 1181–1197. doi:ten.1016/j.cell.2018.02.016. PMID 29522741.
- ^ Cavalier-Smith T (March 2002). "The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa". Int. J. Syst. Evol. Microbiol. 52 (Pt 2): 297–354. doi:10.1099/00207713-52-2-297. PMID 11931142.
- ^ Barry ER, Bong SD (December 2006). "Dna replication in the archaea". Microbiology and Molecular Biology Reviews. 70 (4): 876–87. doi:10.1128/MMBR.00029-06. PMC1698513. PMID 17158702.
- ^ Lane N (2015). The Vital Question – Energy, Evolution, and the Origins of Complex Life . WW Norton. p. 77. ISBN978-0-393-08881-6.
- ^ Forterre P (2006). "Iii RNA cells for ribosomal lineages and three DNA viruses to replicate their genomes: A hypothesis for the origin of cellular domain". PNAS. 103 (10): 3669–3674. Bibcode:2006PNAS..103.3669F. doi:10.1073/pnas.0510333103. PMC1450140. PMID 16505372.
External links [edit]
![]()
Wikimedia Eatables has media related to Procaryota.
- Prokaryote versus eukaryote, BioMineWiki
- The Taxonomic Outline of Leaner and Archaea
- The Prokaryote-Eukaryote Dichotomy: Meanings and Mythology
- Quiz on prokaryote beefcake
- TOLWEB page on Eukaryote-Prokaryote phylogeny
![]() This article incorporates public domain cloth from Science Primer. NCBI. Archived from the original on 2009-12-08.
This article incorporates public domain cloth from Science Primer. NCBI. Archived from the original on 2009-12-08.
Are Prokaryotic Cells In Humans,
Source: https://en.wikipedia.org/wiki/Prokaryote
Posted by: kongnoestringthe.blogspot.com


0 Response to "Are Prokaryotic Cells In Humans"
Post a Comment